Adjusting method of clustering
This tutorial will show you how your results can vary from chosen method of clustering and it also contains tips about adjusting method of clustering to acquire desired results. A 10 ns simulation of murine soluble epoxide hydrolase structure (PDB ID: 1CQZ) was used to run sample calculations and test different clustering methods. Method of clustering in AQUA-DUCT can be easily changed through config.txt file. The inspection of MD simulations suggests that optimal clustering method should group inlets into one main cluster (C1), and two smaller clusters (C2 and C3). Defined clusters with AQUA-DUCT correspond to the hollows in the protein surface (Figure 1b) and groups of entry/exits of smoothed trajectories of water molecules (Figure 1c). Other inlets should be classified as outliers, since transport of solvent molecules through the rest of the tunnels is limited in comparison to the main clusters.
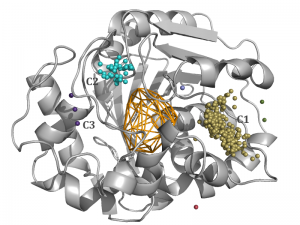
Fig. 1a. Optimal results of clustering of AQUA-DUCT results with meanshift method, bandwidth = Auto and cluster_all = True of 1CQZ with three main clusters provided: C1 (gold), C2 (light blue) and C3 (purple) and other minor 1- or 2-inlet clusters
Please download full tutorial here: TIPS_AND_TRICKS_CLUSTERING
